In order to facilitate and guide clinical interpretation of sequence changes identified from either gene panel, exome or whole genome sequencing, the Personal Cancer Genome Reporter was developed to: i) systematically interrogate cancer genomes in the context of diagnostic, prognostic, and therapeutic biomarkers, ii) prioritize and highlight the most important findings, and iii) present the results in a format accessible to clinical experts.
Comprehensive clinical report from molecular diagnostics in cancer

Cancer is a genetic disease, as DNA is altered in each tumor. The changes differ greatly between different tumors and over time in a tumor. Many changes can occur in each tumor, and these changes may represent targets for therapy, or serve in other ways to inform on tumor management.
The workflow takes DNA sequence variation identified from sequencing (here single nucleotide variants (SNVs), or insertions or deletions (InDels), as well as copy number variant segments of DNA (CNA) as input, and outputs an annotated version using existing knowledge resources.
PCGR integrates a comprehensive set of knowledge resources related to tumor biology and therapeutic biomarkers, both at the gene and variant level.
The application generates a tiered report that will aid the interpretation of individual cancer genomes in a clinical setting, and it builds from publicly available resources to integrate knowledge in a structured way.
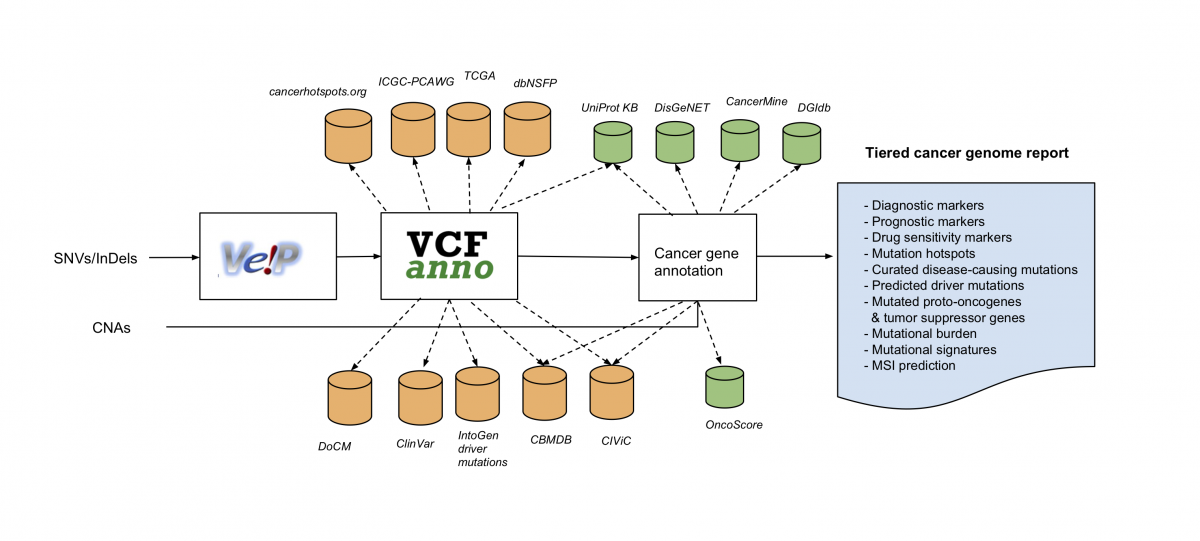
The result is a structured report, here represented by an excerpt of a sample report:

Status:
Finished
Partners:
Oslo University Hospital - Intervention center
Topics
Documentation

Eivind Hovig
Professor and Manager
OUS - Dept. of Tumor Biology and UiO - Dept. of informatics

Relevant Projects


