Monogenic disorders are conditions causally related to changes in a single gene. Several thousand different monogenic disorders are identified to date. Deep insight into the underlying biological mechanisms of disease allows stratification of diseases into subtypes, thus enabling precision medicine. An understanding of the relationship between genotype and phenotype is required. The genotype describes hereditary information, in medical genetics the genotype is defined as genomic variation differing from a reference. The phenotype of a patient, as traditionally perceived, is the constellation of all observed properties deviating from the normal. Phenotypic descriptions in publications and electronic medical records are often imprecise. A precise and comprehensive phenotyping performed in a systematic manner is a prerequisite to enable integration with genomic variation.
Rare genetic variants underlie many congenital presentations. High throughput sequencing has emerged as the preferred method for identifying the molecular genetic etiology of a suspected monogenic disorder. A growing body of evidence supports the use of whole genome- or whole exome sequencing for early molecular diagnoses of neonates and children with life-threatening conditions, entering intensive care treatment. Early diagnoses improve outcomes and reduce costs. When performed on exome or genome scale, the output (number of variants) is vast, from 50 000 variants and upwards in a typical exome, many times multiplied in genome data sets. Hence, predefined areas of interest or prioritization of genetic variants based on deep phenotyping of the patient will effectively reduce workload and increase the diagnostic precision when performing diagnostic high throughput sequencing.
Objectives
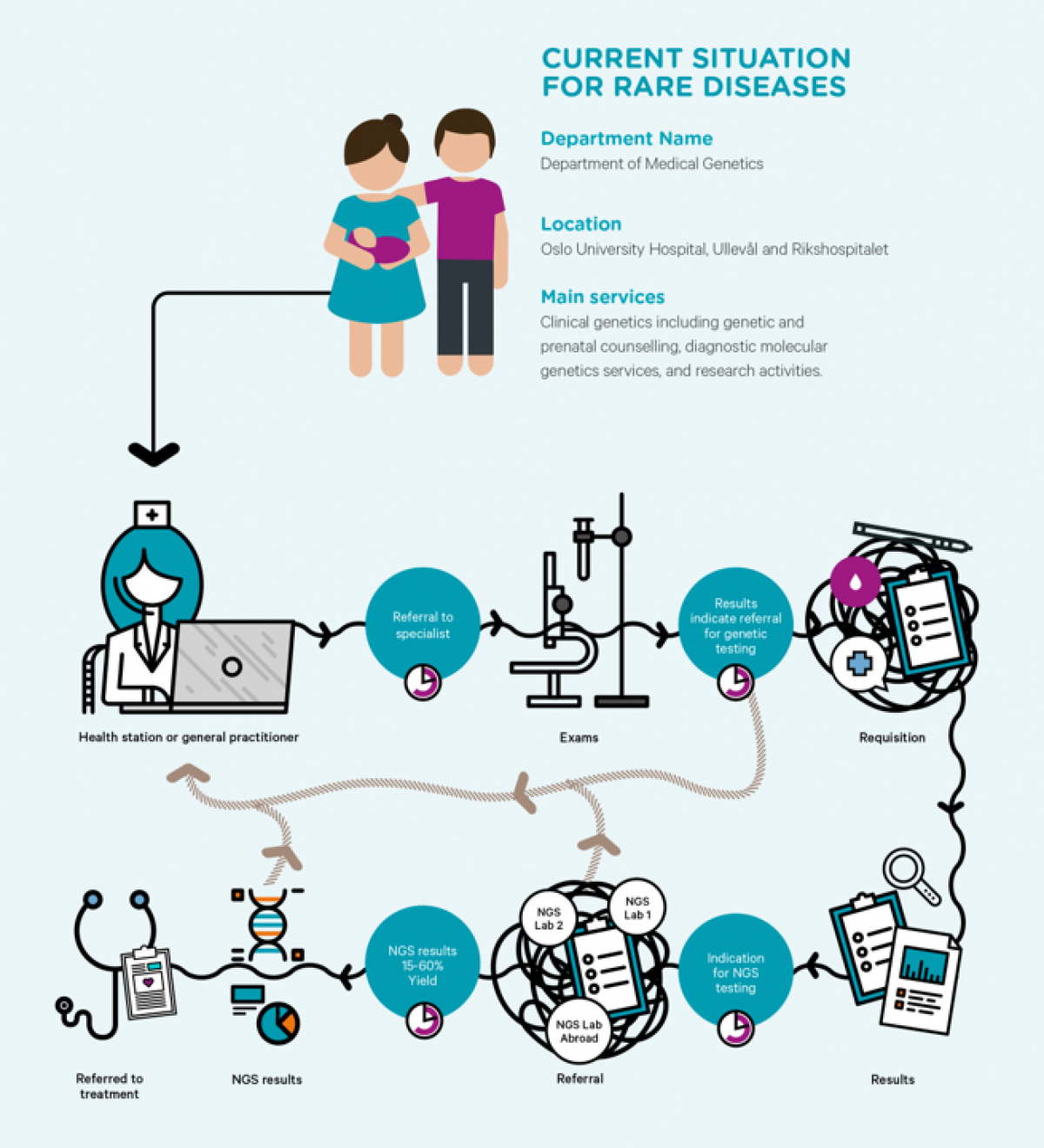
The overall objective of this work package is to promote development of enhanced clinical and laboratory decision support for diagnostics of patients and families with rare diseases, including increased precision and speed of genetic testing.
Tasks
In collaboration with BigMed partners including TSD, the Department of Medical Genetics at OUS is developing and testing informatics solutions for automatized, scalable, high-speed processing of whole genome sequence data.
Front- and backend solutions for systematized patient phenotyping to benefit rapid identification of pathogenic genomic variation are being tested and developed in collaboration with DIPS and IFI, including mapping of ontologies for structured phenotyping.
To facilitate informed consent from families in extremely stressful situations, BigMed partners, including the legal dpt. at OUS and the research dpt. at DNVGL, have mapped needs and requirements for patient consent solutions. TSD has developed and deployed an electronic consent solution based on requirements from the Dpt. of medical genetics, OUS.
Deliverables
Prepare grounds for implementation of whole genome sequencing with low turnaround time, thereby increasing the usefulness of the technology for critically ill patients
Provide recommendations for workflow-solutions
enabling increased precision in analysis and interpretation of genetic variants,
including requisitions and consents.