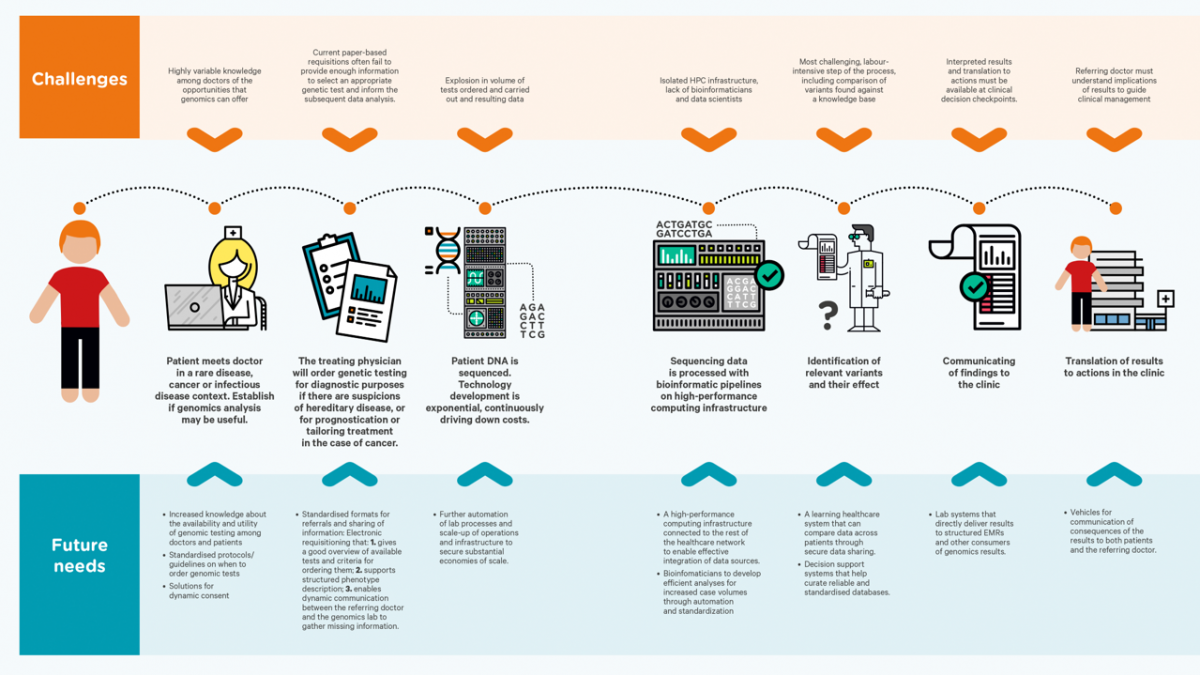
The bioinformatics pipelines currently in use vary greatly from site to site. There is urgent need for standardization of reporting and use of pipeline components, to ensure equal quality of input to genetic clinical decision support systems. Toward this end, BigMed is developing current OUS pipelines, and mapping and benchmarking these to other pipelines, while tackling quality control issues along the entire pipelines, from initial sample to clinical report.
Activities and deliverables are described below.
Somatic sequencing analysis pipelines
- Gene panel DNA sequencing
- Whole exome DNA sequencing
- Somatic RNA-seq analysis pipeline
- Germ-line whole genome sequencing
Quality control
- Benchmarking of pipelines
- Report: Mapping of regulatory frameworks and quality assurance for NGS-based diagnostics
- Report: Framework for quality in NGS
- Report: Mapping of best practice molecular diagnostics in clinical oncology
Clinical reporting
- Genomics-based reports for use with clinical systems.
- Clinical decision support module for tumor board meeting dashboard